The analysis of neuronal structure and its relation to function has become a fundamental pillar of neuroscience since its earliest days, with the underlying premise that morphological properties can modulate neuronal computations. It is often the case that the rich three-dimensional structure of neurons is quantified by tools developed in fields other than neuroscience, such as graph theory or computational geometry; nevertheless, some of the more advanced tools developed in these fields have not yet been made accessible to the neuroscience community. Here we present NCD, Neural Collision Detection, a library providing high-level interfaces to collision-detection routines and alpha-shape calculations, as well as statistical analysis and visualization for 3D objects, with the aim to lower the entry barrier for neuroscientists into these worlds. Our work here also demonstrates a variety of use cases for the library and exemplary analysis and visualization, which were carried out with it on real neuronal and vascular data.
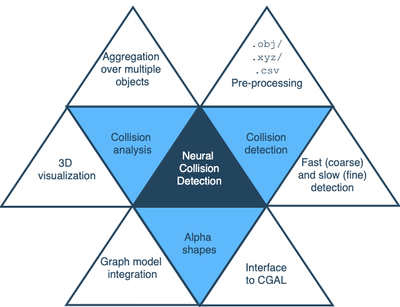
The library at the heart of this work, NCD, uses state-of-the-art computational geometric techniques and algorithms to perform collision detection at scale, with major focus on performance and accuracy. We designed an elaborate pipeline, which starts from the complex components in the neurovascular system—the blood vessels and the neurons in the brain—and finishes with detailed analysis of collision data of the different components, including correlation between the likelihood of a collision at some points on the neuron and geometric properties of these points like graph distance and the critical values of their alpha-shapes.
